Presentation
Rationale
In the field of personalized medicine, the growth of research projects based on extensive data collections of digital images is inevitable and requires a scalable infrastructure for storage, communication, wide distribution, processing and analysis of these data.
Managing large databases of medical images adds a degree of complexity beyond other medical or genomic data due to the size, difficulty of semantic classification, and requirements for appropriate software tools for visualization, analysis and processing of images.
Vision
- Build on the track record of the development of medical imaging software (OsiriX, Weasis and dcm4che) and existing IT infrastructures and scientific expertise in medical informatics at the Geneva Campus Biotech to establish a platform for storage and exchange of medical imaging data suitable for scientific deep learning research projects for AI and personalized medicine.
- Support the development of large open access medical imaging databases based on voluntary patient donation and general consent.
- Should enable the development of individualized and optimized disease management and patient stratification, through the combination of phenotyping (imaging) and genotyping (genetics and biomarkers).
- Will encourage multi-centric clinical studies based on “open clinical data” principles and offer an open repository for multi-modality imaging and ancillary data, genomic profiles, clinical observations and quantitative analysis results.
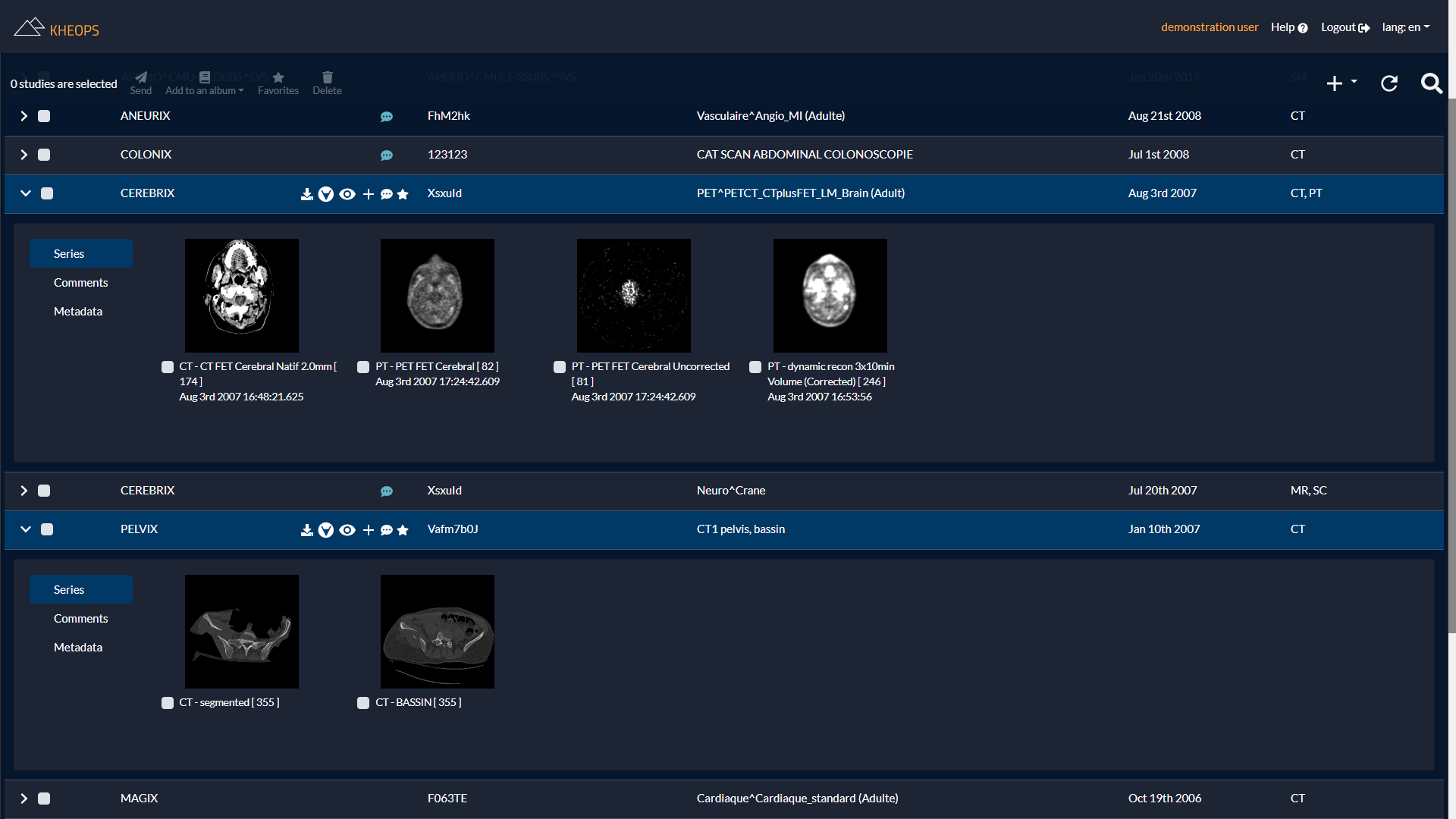
Manage and share your images easily
Once the images have been uploaded (manually or through a DICOM gateway), KHEOPS allows sharing the studies or the series to other users.
The series can be grouped into albums which are themselves easily shareable.
The solution allows visualizing the data from the portal with different viewers: OHIF (HTML5 and mobile), Weasis (multi-platform) and OsiriX (Mac).
Technology
KHEOPS gathers several back-end components developed in Java and with a RESTful API. The storage and the DICOM services are based on the dcm4chee server and the dcm4chee toolkit.
The front-end is developed with the JavaScript framework Vue.
Identity management is managed by Keycloak.
The development and the deployment of the solution follow the DevOps practices (git, travis, sonar, docker-hub).
The solution for monitoring the whole system is Kibana.
The KHEOPS source code is licensed under the MIT license :
A short and simple permissive license with conditions only requiring preservation of copyright and license notices. Licensed works, modifications and larger works may be distributed under different terms and without source code.
Interoperability and open standards
All the data queries and transfers are based on DICOMWeb standard (STOW-RS, QIDO-RS and WADO-RS)
A RESTful API allows interacting with the back-end services.
Authentication is based on OpenID Connect and Keycloak.
KHEOPS NEWS
KHEOPS was at RSNA
At the end of 2019, KHEOPS was ready to be presented at RSNA 2019.
We are very proud of the success and enthusiasm that KHEOPS received during this week.
We hope to be present again at this event, which brings together the radiologist community, to present KHEOPS with its lot of novelties !
The flyer presenting KHEOPS to RSNA is available here.

ABOUT KHEOPS
The KHEOPS solution is developed at Campus Biotech in Geneva. This DICOM-compliant image archive provides a flexible and open image archiving solution for scientists and researchers seeking secure and reliable storage of their data.
Open source project
KHEOPS is an open source project available on GitHub.
All API resources are documented and a user guide is available.






